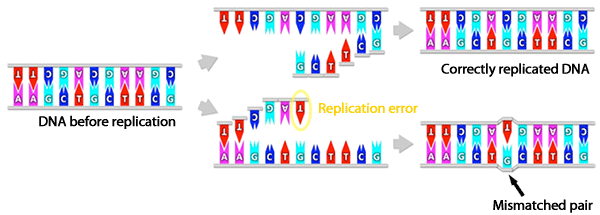
DNA polymerases are the only site where replication errors can actually exist. Most of the errors that do occur are called BASE-PAIR MISMATCHES, which can be corrected by proofreading mechanisms or by a DNA repair mechanism that corrects errors after replication has occurred.
PROOFREADING MECHANISM
The proofreading mechanism depends on the ability for DNA polymerases to back up and remove mispaired nucleotides from a DNA strand. Note that only when the most recently added base is correctly paired with its complementary base on the template strand will the DNA polymerase continue to add nucleotides.
SO HOW DOES IT WORK EXACTLY?
- The correct pairs allow the fully stabilizing hydrogen bonds to form
- If a newly added nucleotide is mismatched, the DNA polymerase reverses using a built in deoxyribonuclease to remove the newly added nucleotide
- The enzymes now resumes moving forward, inserting the correct nucleotide
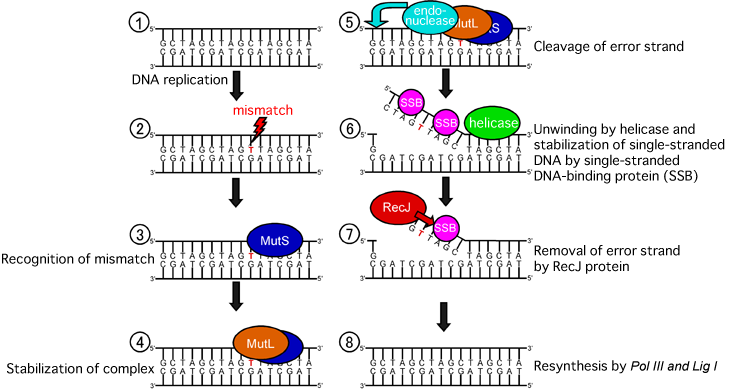
DNA REPAIR MECHANISM
Mispaired bases are either too large or too small to maintain correct separation and results in the non-formation of hydrogen bonds. The base mismatches distort the structure of the DNA helix. These distortions provide the recognition sites for enzymes catalyzing the mismatch pair.
HOW DOES IT WORK?
- The repair enzymes move along the double helix, scanning the DNA for any distortions
- If the enzymes encounter a distortion, they remove a portion of the new chain, including the mismatched nucleotides.
- The gap that is left over is then filled by DNA polymerase using the template stand as a guide
- The repair is completed by DNA ligase which seals the nucleotide chain into a continuous DNA molecule

Deoxyribonuclease I (usually called DNase I), is an endonuclease coded by the human gene DNASE1. DNase I is a nuclease that cleaves DNA preferentially at phosphodiester linkages adjacent to a pyrimidine nucleotide, deoxyribonuclease i
ReplyDelete